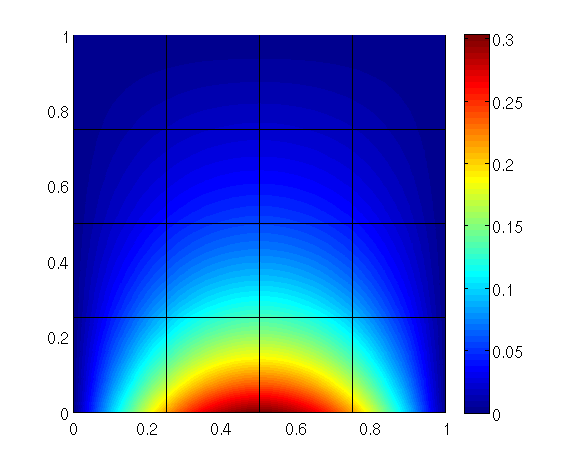
In this tutorial the implementation of inhomogeneous Neumann boundary conditions using trace spaces is shown.
Now we derive the corresponding variational formulation of the introduced problem: find
The mesh is read from three files containing the coordinates, the elements and the boundary attributes of the mesh.
In our example, the edges are given the following attributes: 1 (bottom), 2 (right), 3 (top) and 4 (left).
If there is an error reading those files, then an exception error message is returned.
else, the mesh is plotted using scaling factor of 100, a greyscale of 1.0 and one point per edge.
The homogeneous Dirichlet boundary conditions are set.
and a set of unsigned integers identifying the edges with Neumann boundary conditions is created.
Note, that in our example Neumann boundary conditions are only prescribed at the bottom boundary with attribute 1.
Using the mesh and the homogeneous Dirichlet boundary conditions, the space can be built. We refine the space two times and set the polynomial degree to three. Then the elements of the space are built and the space is plotted.
The right hand side is computed. In our case, the vector rhs only contains the integrals of the Neumann trace as the source term is zero.
We solve the equation using a Mumps solver.
Mesh: Import2dMesh(ncell = 1)
RHS Vector:
Vector(132, [ 1.678744e-01, 0.000000e+00, 1.570060e-02, 5.980456e-03, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 2.374103e-01, 0.000000e+00, 3.790460e-02, 2.477186e-03, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 1.678744e-01, 0.000000e+00, 3.790460e-02, -2.477186e-03, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 1.570060e-02, -5.980456e-03, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00])
System Matrix:
SparseMatrix(132x132, HashedSparseMatrix: 1794 (10.2961%) entries bound.)
Solver:
Mumps(n = 132)
Solution:
Vector(132, [ 2.138851e-01, 9.326650e-02, 3.516756e-02, 5.583589e-03, -4.883840e-02, 3.355068e-03, 1.532811e-02, 2.432564e-03, -8.058410e-03, 5.673075e-04, -1.285410e-03, 9.597194e-05, 3.024792e-01, 1.318988e-01, 8.490199e-02, 2.312798e-03, -6.906793e-02, 4.744783e-03, 3.700534e-02, -1.007601e-03, -1.945472e-02, 1.369602e-03, -5.324341e-04, 3.975288e-05, 5.610624e-02, 3.967310e-02, -2.989946e-02, 2.111419e-03, 1.574144e-02, 4.286670e-04, -2.114211e-02, -1.492999e-03, -8.387079e-03, 5.915169e-04, -2.281498e-04, 1.597013e-05, 6.520320e-03, 1.034894e-03, -3.474042e-03, 2.450143e-04, -5.508022e-04, 3.855529e-05, 2.138851e-01, 9.326650e-02, 8.490199e-02, -2.312798e-03, -4.883840e-02, 3.355068e-03, 3.700534e-02, -1.007601e-03, -1.945472e-02, 1.369602e-03, 5.324341e-04, -3.975288e-05, 3.516756e-02, -5.583589e-03, 1.532811e-02, 2.432564e-03, -8.058410e-03, 5.673075e-04, 1.285410e-03, -9.597194e-05, 3.967310e-02, 6.520320e-03, 1.034894e-03, -2.114211e-02, -1.492999e-03, -3.474042e-03, 2.450143e-04, 5.508022e-04, -3.855529e-05, 1.574144e-02, 4.286670e-04, -8.387079e-03, 5.915169e-04, 2.281498e-04, -1.597013e-05, 1.459270e-02, 2.063719e-02, -8.630098e-03, 6.986304e-04, 5.790065e-03, -1.576721e-04, -1.220480e-02, -9.880126e-04, -3.424278e-03, 2.772246e-04, 9.325616e-05, -7.553547e-06, 2.398323e-03, 3.806541e-04, -1.418383e-03, 1.148302e-04, 2.251403e-04, -1.823587e-05, -2.320734e-03, -4.064907e-04, -3.814143e-04, 6.680687e-05, 6.053633e-05, -1.060309e-05, -3.282014e-03, -5.748646e-04, -9.208157e-04, 1.612860e-04, 2.507497e-05, -4.391942e-06, 1.459270e-02, -8.630098e-03, 6.986304e-04, 2.398323e-03, 3.806541e-04, -1.418383e-03, 1.148302e-04, -2.251403e-04, 1.823587e-05, 5.790065e-03, -1.576721e-04, -3.424278e-03, 2.772246e-04, -9.325616e-05, 7.553547e-06, -2.320734e-03, -4.064907e-04, -9.208157e-04, 1.612860e-04, -2.507497e-05, 4.391942e-06, -3.814143e-04, 6.680687e-05, -6.053633e-05, 1.060309e-05])
1
2 from libconceptspy import concepts
3 from libconceptspy import graphics
4 from libconceptspy import hp1D
5 from libconceptspy import hp2D
6
7 import os
8 from sys import exit
9 from mpi4py import MPI
10
11 SOURCEDIR = os.environ['CONCEPTSPATH' ]
12
13 def main():
14 try :
16 elms = SOURCEDIR + "/applications/unitsquareElements.dat" ,
17 boundary = SOURCEDIR + "/applications/unitsquareEdges.dat"
18 )
19 print ('Mesh:' )
20 print (mesh)
21 except RuntimeError:
22 print("Mesh import error" )
23 else :
24 graphics.MeshEPS_r(msh = mesh, filename = "mesh.eps" , scale=100, greyscale=1.0, nPoints=1)
25
27 for i in range(2,5):
28 boundaryConditions.add(attrib = i, bcObject =
concepts.Boundary (type = concepts.Boundary.DIRICHLET))
29
30
31 NeumannFormula = concepts.ParsedFormula_r(formula = "(sin(pi*x))" )
32 attrNeumann = concepts.Set_uint(1)
33
34
35 levelOfRefinement = 2
36 polynomialDegree = 3
38 p = polynomialDegree, bc = boundaryConditions)
39 space.rebuild()
40 graphics.MeshEPS_r(spc = space, filename = "space.eps" , scale=100, greyscale=1.0, nPoints=1)
41
42
44
45
46 lformNeumann = hp1D.Riesz_r(frm = NeumannFormula)
47 rhs = concepts.Vector_r(spc = tscpNeumann, lf = lformNeumann)
48 print('\nRHS Vector:' )
49 print(rhs)
50
51
52 la = hp2D.Laplace_r()
53 A = concepts.SparseMatrix_r(spc = space, bf = la)
54 A.compress()
55
56 id = hp2D.Identity_r()
57 M = concepts.SparseMatrix_r(spc = space, bf = id)
58 M.compress()
59 M.addInto(A,1.0)
60 print('\nSystem Matrix:' )
61 print(A)
62
63
64 solver = concepts.Mumps_r(A = A)
65 sol = concepts.Vector_r(spc = space)
66 solver(fncY = rhs, fncX = sol)
67 print('\nSolver:' )
68 print(solver)
69 print('\nSolution:' )
70 print(sol)
71
72
73 hp2D.IntegrableQuad.setTensor(concepts.TRAPEZE, True , 8 )
74 space.recomputeShapefunctions()
76
77 if __name__ == "__main__" :
78 main()
![\[ - \Delta u + u = 0 \]](form_185.png)
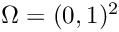
![\[
\nabla u\cdot \underline{n} = g(x,y), \qquad (x,y)\in\Gamma_{\rm N}=\{(x,y)\in\partial\Omega\mid y=0\},
\]](form_745.png)
![\[
u = 0, \qquad (x,y)\in\Gamma_{\rm D}=\partial\Omega\setminus\Gamma_{\rm N},
\]](form_746.png)
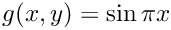
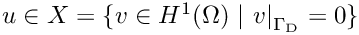
![\[
\int_{\Omega} \nabla u \cdot \nabla v \;{\rm d}x
+ \int_{\Omega} u v \;{\rm d}x
=
\int_{\Gamma_{\rm N}}gv\;{\rm d}x
\qquad\forall v\in X.
\]](form_747.png)