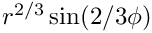Two dimensional FEM in Concepts. This code features high order Ansatz functions and irregular meshes (local, anisotropic refinements in h and p ). All classes for the hp -FEM in two dimensions can be found in the namespace hp2D .
The equation solved is a reaction-diffusion equation (without reaction term)
on the L shaped domain (-1,1)2 \ (0,1)x(-1,0) with mixed boundary conditions chosen such that the exact solution is
The exact solution is
The convergence in energy norm is shown in the results section together with plots of the solution.
Commented Program
FEM Routine
Main Program
Results Complete Source Code
Commented Program
System include files.
#include <memory>
#include <cmath>
#include <cstdlib>
#include <cstring>
#include <fstream>
#include <iostream>
#include <sstream>
#include <unistd.h>
Concepts include files. hp2D.hh is for the high order method.
#include "basics.hh"
#include "toolbox.hh"
#include "formula.hh"
#include "geometry.hh"
#include "space.hh"
#include "function.hh"
#include "graphics.hh"
#include "operator.hh"
#include "hp2D.hh"
With this using directive, we do not need to prepend concepts:: before Real everytime.
The only parameters of the routine fem are some input parameters in input, the boundary conditions in bc and the additional argument output is used to for the gathering the output.
Set up some abbreviations for some parameters found in input. If any of those parameters does not exist in input, an exception is thrown.
l is the number of levels to compute on and p the initial polynomial degree.
{
const uint l = input.
getInt (
"level" );
const uint p = input.
getInt (
"polynomial" );
int getInt(const char *name, const int value=INT_MAX) const
meshNameBase contains the base name of the mesh files.
const std::string meshNameBase = input.
getString (
"meshNameBase" );
std::string getString(const char *name, const char *value=0) const
exenergy gets the value of the exact energy.
const Real exenergy = input.
getDouble (
"exenergy" );
double getDouble(const char *name) const
Returns a double from the hash of doubles.
solverType holds the type of solver to use:
const uint solverType = input.
getInt (
"solver" );
The following lines put together the names for the files which contain the coordinates, the elements and the boundary conditions of the mesh.
std::stringstream fileCoord;
fileCoord << meshNameBase << "Coord.dat" << std::ends;
output.
addString (
"coordFile" , fileCoord.str().c_str());
std::stringstream fileElements;
fileElements << meshNameBase << "Elements.dat" << std::ends;
output.
addString (
"elementsFile" , fileElements.str().c_str());
std::stringstream fileBoundary;
fileBoundary << meshNameBase << "Boundary.dat" << std::ends;
output.
addString (
"boundaryFile" , fileBoundary.str().c_str());
void addString(const char *name, const char *value)
Adds a string to the hash of strings.
Then, these names are given to the mesh importer which reads the files and creates a mesh from it.
fileBoundary.str());
std::cout << "msh = " << msh << std::endl;
Using the mesh, the space can be built. Note that the elements are not yet created as this is an adaptive space. It builds its elements only on request via hp2D::Space::rebuild()
Now, the loop over the different computational levels is initialized.
for (uint i = 0; i < l; ++i) {
std::cout << "i = " << i << std::endl;
timer is used to clock some parts of the code.
Timer and resource monitor.
Now, it is time to build the elements of the space. If there are degrees of freedom in the space, it might be worth to compute something. Otherwise, we skip directly to the refinement of the space .
space.rebuild();
if (space.dim() > 0) {
The time to build the space and the number of degrees of freedom are stored to generate some statistics in the post-processing.
void addArrayDouble(const char *array, const bool newArray=false)
void addArrayInt(const char *array, const bool newArray=false)
The newly built space is printed and written to file in different formats.
std::cout << " space = " << space << std::endl;
MeshDX< Real > drawMeshDX(concepts::Mesh &msh, std::string filename)
MeshEPS< Real > drawMeshEPS(concepts::Mesh &msh, std::string filename, const Real scale=100, const Real greyscale=1.0, const unsigned int nPoints=2)
The right hand side is computed. In our case, the vector rhs only contains the integrals of the Neumann boundary conditions as the right hand side f is 0.
The stiffness matrix is computed from a linear combination of the two bilinear forms hp2D::Laplace and hp2D::Identity . In our case, c is 0 and there is no contribution of hp2D::Identity . Instead of using concepts::BilinearFormLiCo , one could also build a linear combination of the matrices using concepts::LiCo . Sometimes, it is a matter of taste, sometimes a matter of efficiency which one is used.
std::vector< concepts::ElementFormulaContainer<Real> > formulas;
formulas.push_back(a11);
formulas.push_back(a21);
formulas.push_back(a12);
formulas.push_back(a22);
#if 0
#else
#endif
#if 0
lamat_M.addInto(la_M, -1);
cout << "error: " << la_M << endl;
#endif
stiff.compress();
std::cout << " matrix: " << stiff << std::endl;
Set< F > makeSet(uint n, const F &first,...)
The next part is the linear solver. There are quite a few available for Concepts and here is a switch case statement offering a choice among the different solvers (if they are installed – hence the #ifdef statements). The variable solver makes use of the common base class concepts::Operator of all solvers: it is a pointer to the solver to be chosen in the switch clause.
std::unique_ptr<concepts::Operator<Real> > solver(nullptr );
std::unique_ptr<concepts::DiagonalMatrix<Real> > diag(nullptr );
std::unique_ptr<concepts::DiagonalSolver<Real> > precond(nullptr );
#ifdef HAS_PETSC
std::unique_ptr<concepts::PETScMat> stiffP(nullptr );
#endif
switch (solverType) {
case 0:
2*stiff.dimX()+100));
break ;
#ifdef HAS_PETSC
case 1:
break ;
#endif
#ifdef HAS_SuperLU
#endif
#ifdef HAS_UMFPACK
#endif
#ifdef HAS_PARDISO
case 4: solver.reset
break ;
#endif
default :
}
#define conceptsException(exc)
Now, everything is prepared to solve the linear system.
sol is an empty vector to hold the solution of the linear system.
Some immediate statistics of the solver.
std::cout << " solver = " << *solver << std::endl;
<< std::endl;
switch (solverType) {
case 0: {
break ;
}
#ifdef HAS_PETSC
case 1: {
break ;
}
#endif
double getArrayDouble(const char *array, const int number) const
#define conceptsAssert(cond, exc)
}
The energy of the FE solution in sol can be computed by multiplying sol and rhs. It is stored together with the relative error of the FE energy.
const Real FEenergy = sol*rhs;
std::cout << " FE energy = " << std::setprecision(20)
<< FEenergy << std::setprecision(6);
const Real relError = fabs(FEenergy - exenergy)/exenergy;
std::cout << ", rel. energy error = " << std::setprecision(20)
<< relError << std::setprecision(6) << std::endl;
If it was selected to have graphical representation of the solution, the data is written to a file using graphics::drawDataGnuplot .
std::stringstream name;
name << "hpFEM2d-" << i << ".gnuplot" << std::ends;
}
}
bool getBool(const char *name) const
Returns a bool from the hash of bools.
void drawDataGnuplot(T &msh_spc, std::string filename, const S &sol, const concepts::ElementFunction< Real > *fun=0)
If there are further computations (ie. i < l-1), the space is refined using hp2D::APrioriRefinement towards the vertex (and/or edge) carying a singularity. This information has to be given a-priori by the user.
if (i < l-1) {
int pMax[2] = { 1, 1 };
input.
getInt (
"edgRef" ), pMax);
post(refineSpace);
}
}
}
Set up the input parameter space (more in the inputoutput.cc tutorial ).
int main(int argc, char ** argv) {
try {
Parse the input file and extract the variables from it. Mainly the boundary conditions of the problem are relevant from the input file.
inputParser.
parse (SOURCEDIR
"/applications/hpFEM2d.concepts" );
Default values for some parameters:
Number of levels to compute on
void addInt(const char *name, const int value)
Adds an int to the hash of ints.
Initial polynomial degree of the space input.
addInt (
"polynomial" , 1);
Graphics?
void addBool(const char *name, const int value)
Adds a bool to the hash of bools.
Coefficient of the diffusion term in the problem
void addDouble(const char *name, const double value)
Adds a double to the hash of doubles.
Coefficient of the reaction term
Right hand side
Attribute of the vertex which has a singularity
Attribute of the edge which has a singularity (none in this case)
Type of solver to use
Base name of mesh input files input.
addString (
"meshNameBase" , SOURCEDIR
"/applications/lshape2D" );
Title of the problem input.
addString (
"title" ,
"singularity in (0,0) in L shaped domain" );
File name for output data std::stringstream outfile;
outfile << argv[0] << ".out" << std::ends;
input.
addString (
"parameterout" , outfile.str().c_str());
File name for graphics of solution std::stringstream outfileG;
outfileG << argv[0] << ".gnuplot" << std::ends;
input.
addString (
"gnuplot" , outfileG.str().c_str());
output.
addString (
"version" ,
"$Id: hpFEM2d.cc,v 1.7 2005/03/11 21:34:02 kersten Exp $" );
Relative error of solution in energy
FE energy of the solution
Degrees of freedom
Number of iterations in the solver
Time to build the elements of the space in the hp2D::Space::rebuild call
Time to compute the stiffness matrix
Time to solve the linear system These data should be printed in tabular form at the end of the program: each array which should be included in the table has to be added to table.
table.
addMap(concepts::ResultsTable::INT,
"dof" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "energyError" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "feenergy" , output);
table.addMap(concepts::ResultsTable::INT, "iterations" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "space_time" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "stiff_time" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "solve_time" , output);
This parses the command line and adds the parameters to input. How this is done is explained in the inputoutput.cc tutorial .
std::string inputfile;
int opt;
while ((opt = getopt(argc, argv, "-f:l:p:g" )) != EOF)
switch (opt) {
case 'l' : input.
addInt (
"level" , std::atoi(optarg));
break ;
case 'p' : input.
addInt (
"polynomial" , std::atoi(optarg));
break ;
case 'f' : inputfile = std::string(optarg);
inputParser.
parse (inputfile);
break ;
case 'g' : input.
addBool (
"graphics" ,
true );
break ;
default :
std::cout << "Usage: " << argv[0]
<< " [-f FILE] [-l LEVEL] [-p DEGREE] [-d]"
<< std::endl
<< "where" << std::endl
<< " FILE: name of the input file" << std::endl
<< " LEVEL: level of refinement" << std::endl
<< " DEGREE: polynomial degree" << std::endl
<< " -g: graphics of solution" << std::endl
<< "Options given after the input file override the values "
<< "read from the"
<< std::endl << "input file." << std::endl;
std::exit(1);
break ;
}
The boundary conditions are read from the input file into input and have to be converted to concepts::BoundaryConditions . The input file contains two arrays: bctype and bcform with the type of the boundary condition and its formula respectively. There are numberbc entries in both arrays and from this data, a concepts::Boundary is constructed and added to bc.
for (
int i = 1; i <= input.
getInt (
"numberbc" ); ++i) {
}
void add(const Set< Attribute > &attrib, const Boundary &bcObject)
Adds boundary condition for a set of attributes.
boundaryTypes
The different boundary condition types.
int getArrayInt(const char *array, const int number) const
std::string getArrayString(const char *array, const int number) const
The parameters and boundary condition are printed to screen.
std::cout << '[' << argv[0] << "]" << std::endl;
std::cout << "--" << std::endl;
std::cout << "Parameters:" << std::endl
<< " input file = " << inputfile << std::endl
<< input;
std::cout << "--" << std::endl;
std::cout << bc << std::endl;
std::cout << "--" << std::endl;
Now, everything is ready to do the computations.
The input and output data is stored in a file. The table of output data is printed to screen and stored in a file.
std::ofstream* ofs = new std::ofstream(input.getString("gnuplot" ).c_str());
*ofs << std::setprecision(20);
table.
print<concepts::ResultsTable::GNUPLOT>(*ofs);
delete ofs;
std::cout << "--" << std::endl;
std::cout << output << std::endl;
std::cout << table << std::endl;
std::cout << "--" << std::endl
<< "Writing gathered data to disk: "
<< input.getString("parameterout" ) << std::endl;
ofs = new std::ofstream(input.getString("parameterout" ).c_str());
*ofs << "/* program:\t" << argv[0] << std::endl
<< " * command:\t" ;
for (int i = 0; i < argc; ++i)
*ofs << argv[i] << " " ;
*ofs << std::endl
<< " * input file:\t" << inputfile << std::endl;
*ofs << " */" << std::endl << std::setprecision(20) << inputParser;
delete ofs;
}
Finally, exceptions are caught and a sane return value is given back.
std::cout << e << std::endl;
return 1;
}
The input file:
double a 1.0
double c 0.0
string f "(0)"
// overkill solution with 6698
double exenergy 1.8362264461350097
string meshNameBase "lshape2D"
int vtxRef 10
int edgRef 0
int numberbc 8
array int bctype {
1 1
2 2
3 2
4 2
5 2
6 2
7 2
8 1
}
array string bcform {
1 "(0)"
2 "( (1/(1+y*y)^(2/3)) * (sin(2/3*atan(y)) - y*cos(2/3*atan(y)))*2/3 )"
3 "( 2/3/((1+x*x)^(2/3)) * (sin(2/3*atan(1/x)) + x*cos(2/3*atan(1/x))) )"
4 "( 2/3/((1+x*x)^(2/3)) * (sin(2/3*(atan(1/x)+pi)) + x*cos(2/3*(atan(1/x)+pi))) )"
5 "( 2/3/((1+y*y)^(2/3)) * (sin(2/3*(atan(-y)+pi)) + y*cos(2/3*(atan(-y)+pi))) )"
6 "( 2/3/((1+y*y)^(2/3)) * (sin(2/3*(atan(-y)+pi)) + y*cos(2/3*(atan(-y)+pi))) )"
7 "( 2/3/((1+x*x)^(2/3)) * (sin(2/3*(atan(-1/x)+pi)) - x*cos(2/3*(atan(-1/x)+pi))) )"
8 "(0)"
}
Output of the program:
[hpFEM2d]
--
Parameters:
input file =
string author "(empty)"
string comment "(empty)"
string f "(0)"
string gnuplot "hpFEM2d.gnuplot"
string meshNameBase "../../../applications/lshape2D"
string parameterout "hpFEM2d.out"
string title "singularity in (0,0) in L shaped domain"
int edgRef 0
int level 3
int numberbc 8
int polynomial 1
int solver 1
int vtxRef 10
double a 1
double c 0
double exenergy 1.83623
array string bcform {
1 "(0)"
2 "( (1/(1+y*y)^(2/3)) * (sin(2/3*atan(y)) - y*cos(2/3*atan(y)))*2/3 )"
3 "( 2/3/((1+x*x)^(2/3)) * (sin(2/3*atan(1/x)) + x*cos(2/3*atan(1/x))) )"
4 "( 2/3/((1+x*x)^(2/3)) * (sin(2/3*(atan(1/x)+pi)) + x*cos(2/3*(atan(1/x)+pi))) )"
5 "( 2/3/((1+y*y)^(2/3)) * (sin(2/3*(atan(-y)+pi)) + y*cos(2/3*(atan(-y)+pi))) )"
6 "( 2/3/((1+y*y)^(2/3)) * (sin(2/3*(atan(-y)+pi)) + y*cos(2/3*(atan(-y)+pi))) )"
7 "( 2/3/((1+x*x)^(2/3)) * (sin(2/3*(atan(-1/x)+pi)) - x*cos(2/3*(atan(-1/x)+pi))) )"
8 "(0)"
}
array int bctype {
1 1
2 2
3 2
4 2
5 2
6 2
7 2
8 1
}
--
BoundaryConditions(1: Boundary(DIRICHLET, ParsedFormula(0)), 2: Boundary(NEUMANN, ParsedFormula( (1/(1+y*y)^(2/3)) * (sin(2/3*atan(y)) - y*cos(2/3*atan(y)))*2/3 )), 3: Boundary(NEUMANN, ParsedFormula( 2/3/((1+x*x)^(2/3)) * (sin(2/3*atan(1/x)) + x*cos(2/3*atan(1/x))) )), 4: Boundary(NEUMANN, ParsedFormula( 2/3/((1+x*x)^(2/3)) * (sin(2/3*(atan(1/x)+pi)) + x*cos(2/3*(atan(1/x)+pi))) )), 5: Boundary(NEUMANN, ParsedFormula( 2/3/((1+y*y)^(2/3)) * (sin(2/3*(atan(-y)+pi)) + y*cos(2/3*(atan(-y)+pi))) )), 6: Boundary(NEUMANN, ParsedFormula( 2/3/((1+y*y)^(2/3)) * (sin(2/3*(atan(-y)+pi)) + y*cos(2/3*(atan(-y)+pi))) )), 7: Boundary(NEUMANN, ParsedFormula( 2/3/((1+x*x)^(2/3)) * (sin(2/3*(atan(-1/x)+pi)) - x*cos(2/3*(atan(-1/x)+pi))) )), 8: Boundary(DIRICHLET, ParsedFormula(0)), 0: Boundary(FREE, (0)))
--
msh = Import2dMesh(ncell = 3)
i = 0
space = Space(dim = 5, nelm = 3)
matrix: SparseMatrix(5x5, HashedSparseMatrix: 15 (60%) entries bound.)
solver = PETSc(solves PETScMat(5x5), it = 1, ksp = bcgs, pc = ilu)
solve time 0
FE energy = 1.7449022359071539867, rel. energy error = 0.049734721128801930023
i = 1
space = Space(dim = 16, nelm = 12)
matrix: SparseMatrix(16x16, HashedSparseMatrix: 90 (35.1562%) entries bound.)
solver = PETSc(solves PETScMat(16x16), it = 6, ksp = bcgs, pc = ilu)
solve time 0
FE energy = 1.7933874905547884104, rel. energy error = 0.023329887046551971153
i = 2
space = Space(dim = 50, nelm = 21)
matrix: SparseMatrix(50x50, HashedSparseMatrix: 402 (16.08%) entries bound.)
solver = PETSc(solves PETScMat(50x50), it = 11, ksp = bcgs, pc = ilu)
solve time 0
FE energy = 1.817608097337950035, rel. energy error = 0.010139462284866132546
--
string boundaryFile "../../../applications/lshape2DBoundary.dat"
string coordFile "../../../applications/lshape2DCoord.dat"
string elementsFile "../../../applications/lshape2DElements.dat"
array double energyError {
0 0.0497347
1 0.0233299
2 0.0101395
}
array double feenergy {
0 1.7449
1 1.79339
2 1.81761
}
array double solve_time {
0 0
1 0
2 0
}
array double space_time {
0 0
1 0.014998
2 0.043994
}
array double stiff_time {
0 0
1 0
2 0
}
array int dof {
0 5
1 16
2 50
}
array int iterations {
0 1
1 6
2 11
}
ResultsTable(
dof energyError feenergy solve_time space_time stiff_time dof iterations
0 0.0497347 1.7449 0 0 0 5 1
1 0.0233299 1.79339 0 0.014998 0 16 6
2 0.0101395 1.81761 0 0.043994 0 50 11
)
--
Writing gathered data to disk: hpFEM2d.out
The plot of the solution is done with the following Gnuplot script:
@include hpFEM2d-solution.gnuplot
<img src="hpFEM2d-solution.png" width="640" height="480" alt="Plot of solution">
The plot of the solution clearly shows the different elements. They
are not connected as concepts::QuadratureRuleGaussJacobi is used
for the numerical integration. This rule does not have any points on
the boundary of the element. One can also see that there are
<span class="math">p + 2</span> integration points used in each
direction per element. In the terminal and first layers,
<span class="math">p = 1</span> and the rest of the layers (only one
in this plot) have <span class="math">p = mk</span> in layer
<span class="math">k</span> for the <b>linear degree distribution
parameter</b> <span class="math">m = 1</span>. Therefore, the
elements in the second layer show 4 quadrature points in each direction
in the plot.
The plot of the convergence history of the relative energy error is
done with the following Gnuplot script:
@include hpFEM2d-conv.gnuplot
<img src="hpFEM2d-conv.png" width="640" height="480" alt="Plot of solution">
The plot in \f$\log-\sqrt[3]{.}\f$ scale shows that the relativ energy
error behaves like \f$\|e\| \leq c \exp(-b N^{1/3})\f$, where
<span class="math">N</span> is the number of degrees of freedom (ndof).
@section complete Complete Source Code
@author Philipp Frauenfelder, 2004
#include <memory>
#include <cmath>
#include <cstdlib>
#include <cstring>
#include <fstream>
#include <iostream>
#include <sstream>
#include <unistd.h>
#include "basics.hh"
#include "toolbox.hh"
#include "formula.hh"
#include "geometry.hh"
#include "space.hh"
#include "function.hh"
#include "graphics.hh"
#include "operator.hh"
#include "hp2D.hh"
{
const uint l = input.
getInt (
"level" );
const uint p = input.
getInt (
"polynomial" );
const std::string meshNameBase = input.
getString (
"meshNameBase" );
const Real exenergy = input.
getDouble (
"exenergy" );
const uint solverType = input.
getInt (
"solver" );
std::stringstream fileCoord;
fileCoord << meshNameBase << "Coord.dat" << std::ends;
output.
addString (
"coordFile" , fileCoord.str().c_str());
std::stringstream fileElements;
fileElements << meshNameBase << "Elements.dat" << std::ends;
output.
addString (
"elementsFile" , fileElements.str().c_str());
std::stringstream fileBoundary;
fileBoundary << meshNameBase << "Boundary.dat" << std::ends;
output.
addString (
"boundaryFile" , fileBoundary.str().c_str());
fileBoundary.str());
std::cout << "msh = " << msh << std::endl;
for (uint i = 0; i < l; ++i) {
std::cout << "i = " << i << std::endl;
std::cout << " space = " << space << std::endl;
std::vector< concepts::ElementFormulaContainer<Real> > formulas;
formulas.push_back(a11);
formulas.push_back(a21);
formulas.push_back(a12);
formulas.push_back(a22);
#if 0
#else
#endif
#if 0
cout << "error: " << la_M << endl;
#endif
std::cout << " matrix: " << stiff << std::endl;
std::unique_ptr<concepts::Operator<Real> > solver(nullptr );
std::unique_ptr<concepts::DiagonalMatrix<Real> > diag(nullptr );
std::unique_ptr<concepts::DiagonalSolver<Real> > precond(nullptr );
#ifdef HAS_PETSC
std::unique_ptr<concepts::PETScMat> stiffP(nullptr );
#endif
switch (solverType) {
case 0:
break ;
#ifdef HAS_PETSC
case 1:
break ;
#endif
#ifdef HAS_SuperLU
#endif
#ifdef HAS_UMFPACK
#endif
#ifdef HAS_PARDISO
case 4: solver.reset
break ;
#endif
default :
}
(*solver)(rhs, sol);
std::cout << " solver = " << *solver << std::endl;
<< std::endl;
switch (solverType) {
case 0: {
break ;
}
#ifdef HAS_PETSC
case 1: {
break ;
}
#endif
}
const Real FEenergy = sol*rhs;
std::cout << " FE energy = " << std::setprecision(20)
<< FEenergy << std::setprecision(6);
const Real relError = fabs(FEenergy - exenergy)/exenergy;
std::cout << ", rel. energy error = " << std::setprecision(20)
<< relError << std::setprecision(6) << std::endl;
std::stringstream name;
name << "hpFEM2d-" << i << ".gnuplot" << std::ends;
}
}
if (i < l-1) {
int pMax[2] = { 1, 1 };
input.
getInt (
"edgRef" ), pMax);
post(refineSpace);
}
}
}
int main(int argc, char ** argv) {
try {
inputParser.
parse (SOURCEDIR
"/applications/hpFEM2d.concepts" );
input.
addInt (
"polynomial" , 1);
input.
addString (
"meshNameBase" , SOURCEDIR
"/applications/lshape2D" );
input.
addString (
"title" ,
"singularity in (0,0) in L shaped domain" );
std::stringstream outfile;
outfile << argv[0] << ".out" << std::ends;
input.
addString (
"parameterout" , outfile.str().c_str());
std::stringstream outfileG;
outfileG << argv[0] << ".gnuplot" << std::ends;
input.
addString (
"gnuplot" , outfileG.str().c_str());
output.
addString (
"version" ,
"$Id: hpFEM2d.cc,v 1.7 2005/03/11 21:34:02 kersten Exp $" );
table.addMap(concepts::ResultsTable::INT, "dof" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "energyError" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "feenergy" , output);
table.addMap(concepts::ResultsTable::INT, "iterations" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "space_time" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "stiff_time" , output);
table.addMap(concepts::ResultsTable::DOUBLE, "solve_time" , output);
std::string inputfile;
int opt;
while ((opt = getopt(argc, argv, "-f:l:p:g" )) != EOF)
switch (opt) {
case 'l' : input.
addInt (
"level" , std::atoi(optarg));
break ;
case 'p' : input.
addInt (
"polynomial" , std::atoi(optarg));
break ;
case 'f' : inputfile = std::string(optarg);
inputParser.
parse (inputfile);
break ;
case 'g' : input.
addBool (
"graphics" ,
true );
break ;
default :
std::cout << "Usage: " << argv[0]
<< " [-f FILE] [-l LEVEL] [-p DEGREE] [-d]"
<< std::endl
<< "where" << std::endl
<< " FILE: name of the input file" << std::endl
<< " LEVEL: level of refinement" << std::endl
<< " DEGREE: polynomial degree" << std::endl
<< " -g: graphics of solution" << std::endl
<< "Options given after the input file override the values "
<< "read from the"
<< std::endl << "input file." << std::endl;
std::exit(1);
break ;
}
for (
int i = 1; i <= input.
getInt (
"numberbc" ); ++i) {
}
std::cout << '[' << argv[0] << "]" << std::endl;
std::cout << "--" << std::endl;
std::cout << "Parameters:" << std::endl
<< " input file = " << inputfile << std::endl
<< input;
std::cout << "--" << std::endl;
std::cout << bc << std::endl;
std::cout << "--" << std::endl;
fem(input, bc, output);
std::ofstream* ofs = new std::ofstream(input.getString("gnuplot" ).c_str());
*ofs << std::setprecision(20);
table.print<concepts::ResultsTable::GNUPLOT>(*ofs);
delete ofs;
std::cout << "--" << std::endl;
std::cout << output << std::endl;
std::cout << table << std::endl;
std::cout << "--" << std::endl
<< "Writing gathered data to disk: "
<< input.getString("parameterout" ) << std::endl;
ofs = new std::ofstream(input.getString("parameterout" ).c_str());
*ofs << "/* program:\t" << argv[0] << std::endl
<< " * command:\t" ;
for (int i = 0; i < argc; ++i)
*ofs << argv[i] << " " ;
*ofs << std::endl
<< " * input file:\t" << inputfile << std::endl;
*ofs << " */" << std::endl << std::setprecision(20) << inputParser;
delete ofs;
}
std::cout << e << std::endl;
return 1;
}
return 0;
}
virtual const uint dimX() const
void addInto(Matrix< H > &dest, const I fact, const uint rowoffset=0, const uint coloffset=0) const
void compress(Real threshold=EPS)
virtual uint dim() const
Returns the dimension of the space.
![\[ - \Delta u = 0 \]](form_151.png)
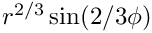
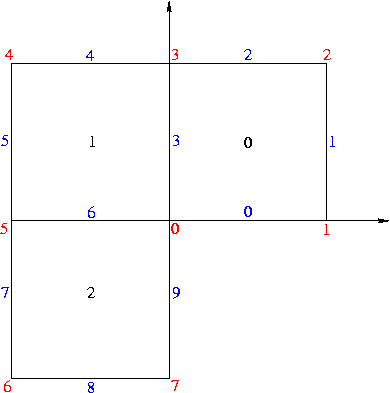 The boundary conditions are homogeneous Dirichlet on the edges 0 and 9 and non-zero Neumann on the rest of the boundary.
The boundary conditions are homogeneous Dirichlet on the edges 0 and 9 and non-zero Neumann on the rest of the boundary.